Strong performance from early models heralds eventual reshaping of care
By Balu Krishnan, PhD, and Andreas Alexopoulos, MD, MPH
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy
Epilepsy is the second most common neurological disorder, impacting 1% to 2% of the world’s population. Individuals with epilepsy typically undergo long-term monitoring of the brain’s electrical activity with EEG recordings for several days. The recorded EEG data are manually reviewed by a trained neurologist, a neurophysiologist or a skilled EEG reader to identify epileptic seizures or interictal discharges that characterize the individual’s epilepsy. Manual review of long-term temporal and spatial EEG data is cumbersome, time-consuming, error-prone and nonscalable, and it may be suboptimal for adequate diagnosis and localization of epilepsy. A scarcity of trained epilepsy subspecialists limits the availability of epilepsy clinics, thereby increasing the cost and delay associated with EEG interpretation and diagnosis.
Computer-aided detection of epileptic seizures has been a research topic within the epilepsy community since the advent of digital EEG. Automatic seizure detection from EEG helps streamline care by promoting rapid and accurate EEG review and interpretation, reducing the need for an expert reviewer and lowering diagnostic and treatment costs.
Advances in deep learning techniques provide new avenues for solving the complex problems inherent in automatic seizure detection. The usefulness of machine learning techniques to detect epileptic lesions on MRI,1 identify the epileptogenic zone2 and classify patients with Rasmussen encephalitis3 has been previously demonstrated by our group. These are just some among many reported applications of artificial intelligence (AI)-mediated diagnostic tools in the management of neurological disorders.
Advertisement
In 2017, Cleveland Clinic’s Epilepsy Center collaborated with Google Inc. to develop an AI system to automatically detect epileptic seizures from long-term EEG recordings in patients undergoing evaluation in Cleveland Clinic’s epilepsy monitoring unit.4 EEG data from approximately 7,000 well-characterized epilepsy patients — corresponding to a total dataset of approximately 20 terabytes — were curated. The capability of an AI system to learn from this rich, unparalleled database can significantly contribute to the clinical care of epilepsy and other neurological disorders. Preliminary evaluation of a subset of data obtained from 1,063 patients (21,655 hours of recording, total of 18,741 seizures) has shown promising results, as we detail below.
Epileptic seizures are characterized by rhythmic electrical oscillation that evolves spatially and temporally. An appropriate neural network for seizure detection should be able to identify the spatiotemporal evolution of epileptic seizures. The temporal graph convolutional network (TGCN) is a deep learning model that leverages spatial information in structural time series (Figure 1). TGCNs extract features that are localized and shared over both temporal and spatial dimensions of the input. TGCNs have built-in invariance to when and where a pattern of interest occurs and thus provide an inductive bias to discriminate these patterns. The TGCN model was used for this purpose in the study we profile below.
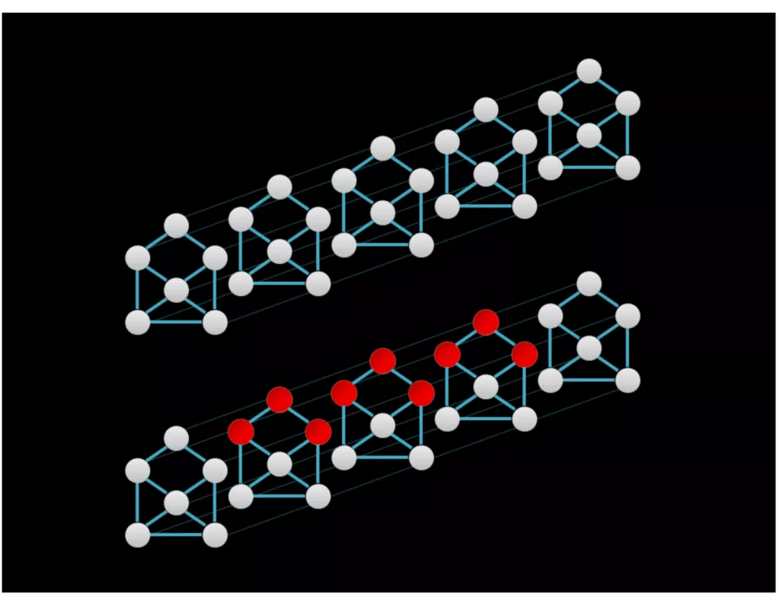
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/a3d43788-e277-442c-a498-e6285ec82fdc/Fig-1-1024x800_png)
Figure 1. Graphical representations of a structural time series with six sequences. Graph structure is depicted by solid lines, whereas temporal adjacency is denoted by the fainter dashed lines. The red nodes in the bottom series indicate receptive fields of feature extraction operation in the temporal graph convolutional network (TGCN), which extracts features that are spatially and temporally localized.
Advertisement
Scalp EEG data from 1,054 patients were used for the study. EEG data were collected with a Nihon Kohden system using a standard 10-20 montage and sampled at 200 Hz. Two sets of data were used:
The temporal location of epileptic events — such as seizures and interictal abnormalities — was annotated by a trained epileptologist. EEG time series were segmented into 96-second epochs and annotated as “seizure” and “seizure-free” epochs. Data segments with seizures were deemed positive, and those without seizures were deemed negative.
Training dataset. The TGCN model was trained using EEG data from 995 patients — clipped data from 613 and continuous EEG data from 382. Approximately 15,000 hours of data were used for training the model. The sparse nature of epileptic seizures led to class imbalance since seizure-free epochs are more frequent than seizure epochs. Data epochs without seizures were subsampled at 90% to balance the two classes.
Tuning and testing datasets. Hyperparameters associated with the neural network architecture were optimized using a tuning set consisting of 30 patients (2,800 hours of recording). Performance of the classifier was evaluated using a testing set consisting of 38 patients (4,000 hours of recording). Only long-term data were used for tuning and testing the deep learning models.
Advertisement
As a preprocessing step, short-time Fourier transform was used to extract informative time frequency features associated with an epileptic seizure (Figure 2).
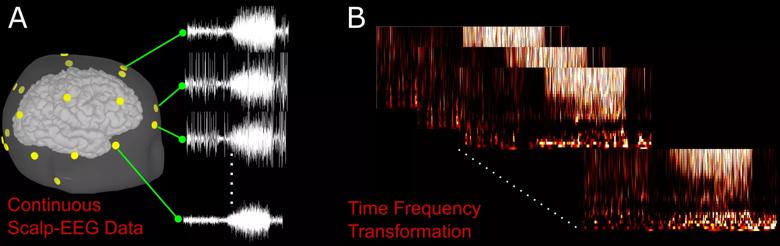
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/6b4d3777-48f2-4a38-906a-85cb7545ff0c/Krishnan-Figure-2_jpg)
Figure 2. (A) Example of a seizure on an EEG from a single patient. Yellow nodes correspond to the location of EEG sensors on the patient’s scalp. (B) Preprocessing of a raw EEG waveform using time frequency transformation.
The neural network architecture consists of a series of spatiotemporal convolutional layers with max pooling along the temporal dimension between the blocks of convolutional layers4 (Figure 3). Five different TGCN architecture configurations were trained and compared, as we have detailed elsewhere.4
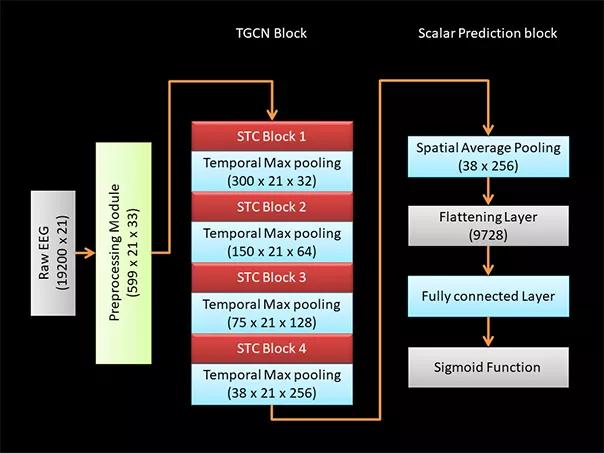
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/145bff0f-0903-43e7-81eb-1bc121e20d20/Krishnan-Fig-3_jpg)
Figure 3. Configuration of TGCN architecture. Dimensions of data at each processing step are indicated in parentheses. A raw EEG waveform of dimensions 19,200 × 21 (time × number of EEG leads) is decomposed using a short-time Fourier transform module to extract the time frequency characteristics of the EEG data. The time frequency decomposed signal has dimensions of 599 × 21 x 33 (time × number of EEG leads × frequency). Four blocks of spatiotemporal convolutional (STC) layers extract localized features over both spatial and temporal dimensions. Temporal max pooling is used to leverage information across a wider time window. To achieve a scalar prediction, the output from the TGCN block is spatially averaged using a pooling layer and flattened. Fully connected layers are used to learn the high-level features, and the scalar prediction is achieved using a sigmoid function.
Advertisement
To emulate real-world scenarios for an epileptic seizure alarm system, model performance was evaluated using (1) sensitivity at 95% specificity, (2) sensitivity at 99% specificity and (3) area under the receiver operating characteristic curve (AUROC). A low number of false positives improves the reliability of the seizure detection tool. Performance of the five TGCN configurations was evaluated using the tuning set, and the TGCN configuration exhibiting superior performance was selected.
The selected TGCN configuration had an AUROC of 0.97, 87% sensitivity at 97% specificity, and 75% sensitivity at 99% specificity. Performance of the TGCN was also compared with other deep learning models that leverage spatiotemporal information for making predictions. The five models tested in the study had performance similar to that of the TGCN.
Overall, on the testing dataset the TGCN achieved an AUROC of 0.93, 64% sensitivity at 97% specificity, and 47% sensitivity at 99% specificity.
One of the major factors limiting adoption of deep learning-based diagnostic tools in healthcare is the lack of transparency of AI systems. It is critical for clinicians to interpret how a deep learning system has made a prediction. Model explainability has significant implications in real-world deployment of deep learning-based diagnostic tools. Additionally, model explainability can provide novel insights into disease diagnosis, providing new research avenues and treatment paradigms. Two separate strategies for model explainability were investigated in this study to extract rich contextual information pertaining to the precise time a seizure occurred and the spatial location of electrodes involved during seizure onset.
Input attribution was the first model explainability tool used in the study. Input attribution assigns an importance level to different input features. Figure 4 shows the input attribution overlaid on an EEG recorded during a single seizure event in a patient. The seizure can be clinically described as follows: Patient has epileptic arousal around 40 to 45 seconds into the EEG data sample, followed by prominent high-frequency activity in the left hemisphere evolving into high-amplitude sharp wave in the Fp1, F7 and F3 leads. The input attribution map corroborates this description, especially through its emphasis on the activity beginning at 40 seconds and the subsequent emphasis on high-frequency activity in the left part of the brain.
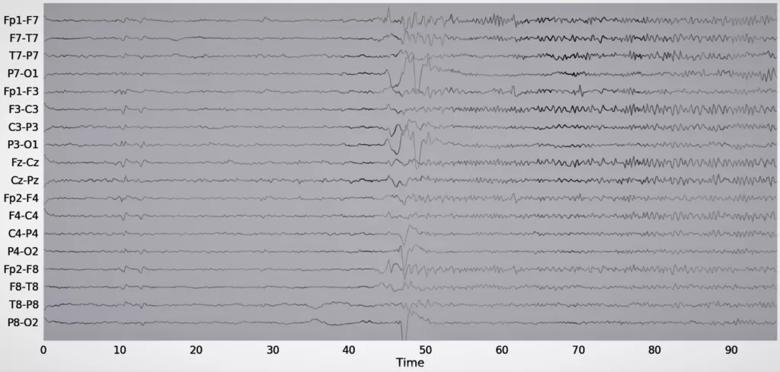
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/af4e9c3a-d7ff-4a31-99b9-12997a855b93/New-Fig-4_resupplied-11-21-19-1024x488_png)
Figure 4. Example of an input attribution map overlaid on EEG data. The attribution score is proportional to the waveform intensity. Reprinted, with permission, from Covert et al.4
Sequence dropout is another strategy of model explainability. In sequence dropout, sets of leads are removed from the model and the model’s performance without those leads is evaluated. Sequence dropout can be assessed by ignoring one lead at a time or by simultaneous removal of multiple leads from part of the brain. Figure 5 is an example of seizure localization using sequence dropout. Sequence dropout identified electrodes in the right frontotemporal region concordant with the clinically identified seizure onset.
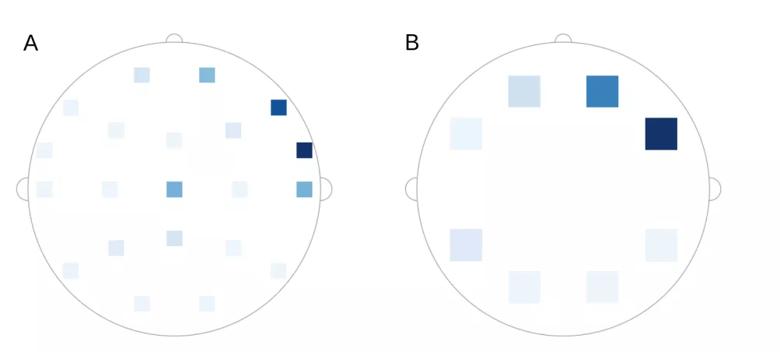
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/d415a097-ed29-44e2-8b55-4ed0fd4330b6/Fig-5-supplied-10-16-19_png)
Figure 5. Example of seizure localization using sequence dropout. Intensity correlates to the reduction in the model’s prediction when (A) one lead was dropped at a time and (B) a set of leads was dropped at the same time. Reprinted, with permission, from Covert et al.4
Our investigations reveal that deep learning models can achieve strong performance in automated detection of epileptic seizures. The performance of the TGCN could be improved by integrating recurrent neural networks, managing class imbalance, ensuring superior labeling and data cleanup, and training on the larger set of curated data. The model’s capability to extract rich contextual information pertaining to seizure onset and spatial localization can further enhance its applicability in real-time automated seizure detection, thereby aiding clinical diagnosis and management.
Finally, electrophysiological information from EEGs could be integrated with results from other epilepsy diagnostic modalities — such as MRI, PET, ictal SPECT, magnetoencephalography and genetic data — along with patient demographics and clinical history to develop an AI-based epilepsy management and diagnosis tool to detect epileptic seizures, recommend appropriate plans of care and optimize patient management. Such tools have great potential to streamline epilepsy care, improve patient access and reduce cost associated with epilepsy management. The capability of AI systems to learn from a wealth of clinical data could be successfully harnessed in future studies that may facilitate medication management, optimize patients’ stay in the epilepsy monitoring unit, detect and potentially predict occurrence of epileptic seizures, identify appropriate candidates for resective epilepsy surgery and predict postoperative outcomes.
Dr. Krishnan is a staff research scientist and Dr. Alexopoulos is a staff physician, both in Cleveland Clinic’s Epilepsy Center.
Advertisement

Aim is for use with clinician oversight to make screening safer and more efficient

Rapid innovation is shaping the deep brain stimulation landscape

Study shows short-term behavioral training can yield objective and subjective gains

How we’re efficiently educating patients and care partners about treatment goals, logistics, risks and benefits

An expert’s take on evolving challenges, treatments and responsibilities through early adulthood

Comorbidities and medical complexity underlie far more deaths than SUDEP does

Novel Cleveland Clinic project is fueled by a $1 million NIH grant

Tool helps patients understand when to ask for help