Screening assay effective across all stages, can identify tissue of origin

Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/5bb6d305-533b-4b95-8fd1-2c661e58a353/HRC_2881047_04-27-22_088_MLC-jpg)
bloodtest_650x450
A prototype blood-based screening test evaluated by Cleveland Clinic researchers and other experts can accurately identify the presence and originating site of 12 high-mortality cancer types, across all stages of progression, with a low false-positive rate, according to the latest results of an ongoing national clinical trial.
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy
The screening test’s sensitivity (its probability of detection or true-positive rate) was 67.3% for stages I-III of the 12 cancer types: anus, bladder, colon/rectum, esophagus, head and neck, liver/bile duct, lung, lymphoma, ovary, pancreas, stomach and plasma cell neoplasm. Together, those cancers account for more than 63% of all U.S. cancer deaths. The test’s sensitivity increased with higher-stage malignancies, ranging from 39% in stage I to 92% in stage IV among the 12 pre-specified cancers.
Its specificity was 99.3%, meaning a 0.7% false positive rate — less than 1% of individuals without cancer would be wrongly identified as having cancer. In the 96% of cases where the test was able to predict the tissue where the malignancy originated, its accuracy was 93%.
Those promising findings raise hopes that the assay will help achieve the long-sought goal of population-scale, broad-based, early detection of cancer in asymptomatic patients. An effective multi-cancer screening test — particularly one that can detect lethal cancer types for which no current screening paradigm exists — could transform oncology care, improving survival chances and treatment outcomes.
At present, only four single-cancer screening regimens exist: the prostate-specific antigen test, mammography, colonoscopy, and computerized tomography to screen patients at high risk for lung cancer. Each has inherent issues and limitations, including cost, access utilization and invasiveness.
“The goal of screening is to cure more cancer and prevent deaths,” says Eric Klein, MD, Chairman of Cleveland Clinic’s Glickman Urological & Kidney Institute, a senior author of the Annals of Oncology report and the co-principal investigator of Cleveland Clinic’s portion of the study along with Taussig Cancer Institute oncologist and Vice Chair for Research Mikkael Sekeres, MD. “Having a blood test is a simpler approach and likely would be met with a high degree of patient acceptance.”
Advertisement
The multi-cancer assay’s preliminary results are “positive in a way that I think people hoped for but were surprised by,” Dr. Klein says. “Although it’s a first-generation test, I think it sets the paradigm for liquid biopsy as a screening tool for cancers.”
Dr. Sekeres adds: “The study’s findings also have the potential to impact how we monitor patients with an established cancer diagnosis — using a simple blood test, instead of the need for multiple scans or repeated biopsies — and have implications for screening patients for cancer across the entire population.”
Annals of Oncology Editor-in-Chief Professor Fabrice André, Director of Research at France’s Institut Gustave Roussy, says: “This is a landmark study and a first step toward the development of easy-to-perform screening tools. Earlier detection of more than 50% of cancers could save millions of lives every year worldwide and could dramatically reduce morbidity induced by aggressive treatments.”
Unlike traditional biopsies, which analyze tissue from a single organ or tissue site to detect a single cancer type, a liquid biopsy in principle can sample the entire body for multiple malignancy types by looking for cancer biomarkers circulating in blood.
The new screening test targets circulating cell-free DNA (cfDNA), which are short, nonencapsulated nucleic DNA fragments deposited in the bloodstream as a consequence of cellular necrosis, apoptosis, secretion or other processes. In a person with cancer, a small portion of the plasma cfDNA load is circulating tumor DNA (ctDNA), originating from the primary tumor or circulating tumor cells and carrying the same molecular aberrations as the source tumor, including genetic mutations and epigenetic modifications.
Advertisement
The test utilizes next-generation deep sequencing and a custom hybridization panel to determine the presence or absence of cancer by obtaining cfDNA’s methylation state and characteristics. Unique methylation patterns of key genes involved in oncogenesis are indicative of specific cancer cell types and the tissue where the tumor originates. These methylomic signatures enables simultaneous detection and localization with a single test.
The assay was developed by GRAIL, Inc., a California-based healthcare company backed by prominent investors including Microsoft co-founder Bill Gates and Amazon founder Jeff Bezos. The test is being evaluated in a multi-site, three-phase case-control observational clinical trial known as the Circulating Cell-free Genome Atlas (CCGA), funded by GRAIL. Cleveland Clinic is one of the CCGA’s 142 study locations and enrolled the largest number of patients of any site — more than 1,500.
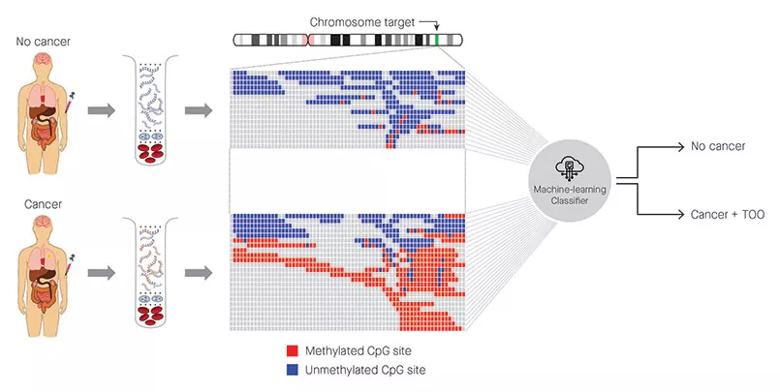
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/427235c0-e188-4f5e-81b5-4a134281a9ed/805x-Inset-Cancer-Screen-Test_jpg)
Figure: Identification of cancer status for more than 50 cancer types, as well as tissue of origin localization, from a single blood draw. Cell-free DNA is isolated from blood samples drawn from a patient without cancer (top) or with cancer (bottom) and subjected to a targeted methylation sequencing assay. Sequencing results identifying methylated (red) or unmethylated (blue) CpG regions are fed into a machine-learning classifier that can identify the presence or absence of cancer, as well as identify the tissue of origin.
Credit: Allen McCrodden, Associate Director, Creative Group, ProEd Communications.
Advertisement
The liquid biopsy approach utilizing cfDNA sequencing and analysis has shown potential in post-diagnostic cancer clinical applications, such as guiding therapy selection and indicating tumor burden, treatment response, resistance and impending relapse. But there had been skepticism about cfDNA’s utility for detecting cancers, particularly at the early stage of oncogenesis when only extremely small amounts of ctDNA are present in plasma.
Since cancer initiation and progression are regulated by both genetic (DNA sequence alterations) and epigenetic (alterations in gene expression and activity) events, there was also uncertainty about where to focus the search for a reliable multi-cancer screening biomarker.
The CCGA’s initial discovery phase sub-study evaluated three potential screening methods to characterize cancer-specific plasma cfDNA signals:
Normally, DNA methylation helps regulate gene expression and stable gene silencing. As research from the Cancer Genome Atlas Program has shown, certain patterns of densely clustered methylation alterations of DNA sequences involved in transcription initiation — specifically, hypermethylation of certain 5’-C-phosphate-G-3’ sites, or CpG islands — are indicative of cancer, and of individual tumor types.
Advertisement
The CCGA discovery-phase sub-study found that the methylation-based assay outperformed the whole-genome and targeted sequencing approaches for multi-cancer detection across stages, with high specificity. That superiority likely is due to methylation’s pervasiveness as a potential signal compared with the mutation sites that other liquid biopsy tests typically sample. Genetic mutation-based screening tests also can be confounded by highly prevalent mutations resulting from biological processes, such as clonal hematopoiesis of indeterminate potential.
“It turned out that, for a variety of reasons, methylation was the best,” Dr. Klein says. “All three methodologies had roughly the same detection rate, but the practical aspects of targeted methylation detection are that it’s cheaper and less complicated to do. And you can develop individual methylation assays for each type of tumor.”
With methylation identified as the preferred basis for the screening assay, the second (current) CCGA sub-study sought to train and validate a machine-learning classifier (an algorithm) to distinguish cancer versus non-cancer and the origin site of any detected malignancy from a plasma specimen. The classifier was used to predict cancer presence and location based on methylation patterns in ctDNA.
The sub-study’s 6,689 participants were divided into a cancer/non-cancer training set and an independent validation set. The training and validation cohorts were generally comparable in demographics. The participants with cancer represented more than 50 primary cancer types across all clinical stages. The large non-cancer cohort included significant numbers of patients with potentially confounding conditions, allowing the researchers to gauge the assay’s specificity in population-level-like screening conditions.
Whole-genome bisulfite sequencing of plasma cfDNA produced 3,508 analyzable samples. Using those results and methylation array data from the Cancer Genome Atlas, the CCGA researchers identified regions of the Genome Reference Consortium human reference genome GRCh37 (hg19) expected to contain cancer- and/or tissue-specific methylation patterns. With training, the classifier derived the most informative targets, ultimately allowing the investigators to create a targeted methylation panel that covered 103,456 genomic regions and 1.1 million CpGs. Methylation patterns determined a sample’s cancer/non-cancer status and the tissue of origin of a malignancy, if detected.
To be effective as a population-level cancer screening test, a candidate assay should be:
The targeted methylation-based multi-cancer test meets those criteria, the CCGA investigators say.
“The really exciting part is that not only do we see positive detection results in cancers for which there is no screening paradigm, but there’s a low false-positive rate and highly accurate predictions for what organ the tumor is located in in those who have a positive test,” Dr. Klein says.
In pancreatic cancer, for instance — which accounts for 7% of all cancer deaths, is usually diagnosed at an advanced stage and lacks a screening test — the assay’s sensitivity was 63% in stage I, 83% in stage II, 75% in stage III and 100% in stage IV.
While a screening test’s sensitivity is important for an individual subject, its specificity is the more relevant metric for widespread use, Dr. Klein says, “At a population level, it’s more important to find as many cancers as are prevalent. Even if a screening test detects only 50% of early-stage cancers, that’s still 50% that aren’t found now.”
A prospective trial using an asymptomatic cohort is needed to precisely calculate the test’s positive predictive value (PPV, the probability that a positive result is indicative of actual disease). But if a similarly performing assay was applied to a population with an annual cancer incidence of 1.3%, the CCGA researchers calculated that the test would identify 715 cancers per 100,000 screened persons and would yield 691 false positive results requiring diagnostic work-ups to rule out disease: a PPV of 51%.
By comparison, U.S. Preventive Services Task Force-recommended screening tests for breast, colorectal and lung cancer have PPVs from 3.7%-4.4%.
Additional studies are needed to determine how well the new test performs in an asymptomatic screening population and whether its clinical use reduces cancer mortality. Other limitations of the current study are that, at the time of the analysis, not all patients had been followed for a year, which is needed to ensure their non-cancer status was accurate; and that some inaccuracy occurred in the detection of tissue of origin for cancers driven by the human papilloma virus.
GRAIL is conducting several research projects to further validate the multi-cancer detection approach. They include a third CCGA sub-study and an observational trial known as PATHFINDER that will assess the test’s use in clinical practice, to determine its impact on diagnostic resolution after detection of a cancer signal and how the results affect patients. Cleveland Clinic plans to participate in PATHFINDER later in 2020.
“One of PATHFINDER’s goals of is to figure out if the test accelerates the diagnosis of cancer in patients who are screened,” Dr. Klein says. “The other thing we’re going to be looking at is physician behavior. What happens after a test is positive? What does the physician do? And what is the patient’s reaction to having a test, including those who receive a negative result? In addition, we will follow patients for a year to determine if the negative tests miss anything.”
With those data and other independent validation results, the company can then make its case for Food and Drug Administration approval of the screening test.
Disclosure: Dr. Klein is a consultant for GRAIL.
Advertisement
Combination therapy improves outcomes, but lobular patients still do worse overall than ductal counterparts
Bringing empathy and evidence-based practice to addiction medicine
Supplemental screening for dense breasts
Combining advanced imaging with targeted therapy in prostate cancer and neuroendocrine tumors
Early results show strong clinical benefit rates
The shifting role of cell therapy and steroids in the relapsed/refractory setting
Radiation therapy helped shrink hand nodules and improve functionality
Standard of care is linked to better outcomes, but disease recurrence and other risk factors often drive alternative approaches