Computer-assisted labeling method offers benefits in clinical, research and training settings
A semi-automated anatomical labeling method for stereotactic electroencephalography (SEEG) contacts demonstrates shorter analysis time and better labeling consistency while reducing the likelihood of gross anatomical error compared with the conventional manual method of brain mapping.
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy
Details of the process and results of an analysis with over 17,000 SEEG contacts were published online May 6 in Epilepsia Open.
“A computerized approach can precisely and consistently label brain anatomy,” says the study’s lead author, Kenneth Taylor, a research associate with Cleveland Clinic’s Epilepsy Center. “The semi-automated method we developed compared favorably with our regular process in terms of accuracy while also saving a great deal of time.”
Implanting depth electrodes under stereotactic guidance allows recording of ictal and interictal intracranial EEG from cortical and subcortical structures. But accurately correlating the electrical activity with anatomy is complicated by various factors, including individual variation in anatomy and abnormalities in brain function or structure resulting in unpredictable epileptic activity. Electrodes can be placed in a variety of trajectories, and the individual contacts may border two or more regions. Moreover, different physicians may use varying terminology when referencing the same areas.
Data were analyzed from more than 17,000 SEEG electrode contacts from 103 patients who underwent SEEG during surgical evaluation for medically intractable focal epilepsy at Cleveland Clinic from 2016 to 2017. At that time, the labels were assigned by trained clinical epilepsy fellows during standard clinical work (“manual method”), a method that typically takes three to four hours per patient.
For the semi-automated method, the normal clinical approach was used through the marking of locations of contacts in the brain. At that point, labels were automatically assigned by correlating the subject’s preoperative MRI with the USCBrain atlas using BrainSuite software. The USCBrain atlas has a total of 130 cortical and 29 noncortical regions and contains 26 sulcus regions per hemisphere, including areas known to be important in patients with medically intractable focal epilepsy.
Advertisement
Results of the two methods were retrospectively compared and found to be in agreement in 82% of contacts (P = 0.852) (Figure). Agreement was comparable regardless of whether patients had a previous surgery. Most patients had 80% to 90% agreement between the methods, and none had less than 70% agreement.
Subsequently, two experts reviewed the labels for which the two methods disagreed and determined that the semi-automated method was more accurate than the manual method (48% vs. 36%).
Next, for labels that the experts disagreed with, the disagreement was categorized as being either in a region adjacent to the correct contact location or a gross error. For the semi-automated method, only 1 of 62 label disagreements was deemed a gross error compared with 5 of 77 label disagreements with the manual method.
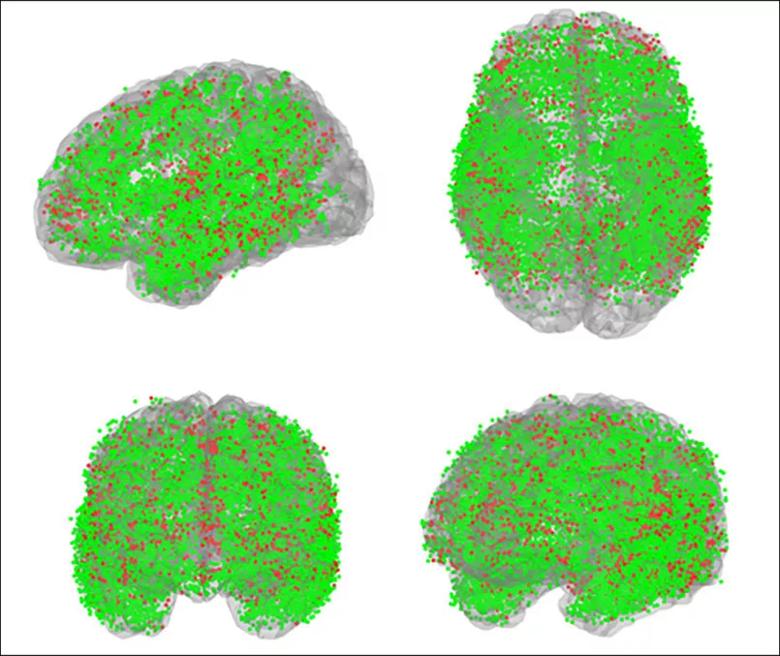
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/72319c59-a95d-49bb-b3ef-e7c0d949c883/21-NEU-2183759_brain-mapping-epilepsy_805x677_jpg)
Figure. More than 17,000 SEEG contacts from 103 Cleveland Clinic patients used to validate the labeling method are shown here in common space from various orientations. Green spheres denote agreement between the manual and automated methods (82% of contacts); red spheres denote disagreement (18%). Expert review favored the automated label more frequently than the manually assigned one.
The authors highlight several advantages of the semi-automated approach to SEEG contact labeling:
Advertisement
“The ability to determine the anatomic location of SEEG electrodes with high accuracy is critically important for diagnosing intractable epilepsy,” adds the study’s senior author, Dileep Nair, MD, Section Head of Adult Epilepsy and Director of Intraoperative Neurophysiologic Monitoring at Cleveland Clinic. “This method can also serve as a useful tool for trainees as they learn SEEG interpretation.”
This research is part of ongoing investigations by this team to enhance brain mapping. They developed a cortico-cortical evoked potentials (CCEPs) technique using low-frequency electrical stimulation of electrodes implanted in patients undergoing invasive monitoring for epilepsy surgery. They used it to map areas and identify functional and epileptic networks, and are developing a single brain atlas using data from hundreds of patients, as summarized in a prior Consult QD post.
In addition, the team developed a method known as the “FAST graph” to more easily visualize the complex data obtained from CCEPs from multiple subjects. It was described last year in Epilepsy Research (2020 Mar;161:106264).
“We are continuing to explore computerization of mapping and labeling, which is a natural fit for something as complex and amenable to data analysis as brain anatomy,” says Dr. Nair. “We expect automated systems to become a normal part of clinical workflow and to more efficiently drive research forward.”
Advertisement
Advertisement

Aim is for use with clinician oversight to make screening safer and more efficient

Rapid innovation is shaping the deep brain stimulation landscape

Study shows short-term behavioral training can yield objective and subjective gains

How we’re efficiently educating patients and care partners about treatment goals, logistics, risks and benefits

An expert’s take on evolving challenges, treatments and responsibilities through early adulthood

Comorbidities and medical complexity underlie far more deaths than SUDEP does

Novel Cleveland Clinic project is fueled by a $1 million NIH grant

Tool helps patients understand when to ask for help