Looking at 3-D modeling techniques to detect differences in PTEN variants
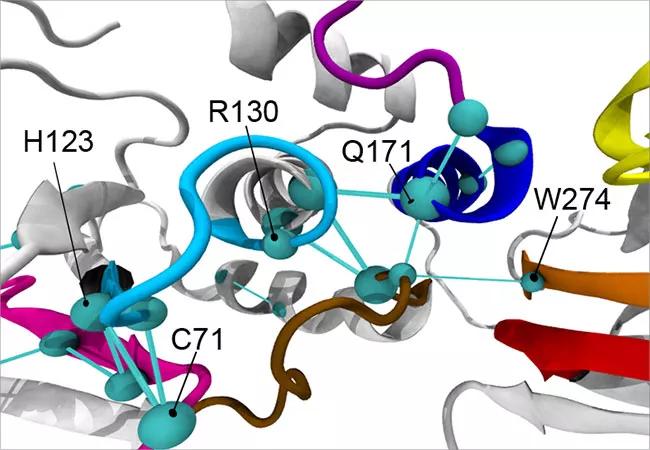
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/19a196b2-ad33-41e1-9e11-2f0f08888418/PTEN-flip-Lerner_jpg)
PTEN flip-Lerner
Germline mutations of the tumor suppressor gene PTEN are associated with autism spectrum disorder (ASD) and Cowden and other syndromes that predispose patients to a broad range of cancers. Charis Eng, MD, PhD, Genomic Medicine Institute, has dedicated much of her career to uncovering and investigating these genetic links.
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy
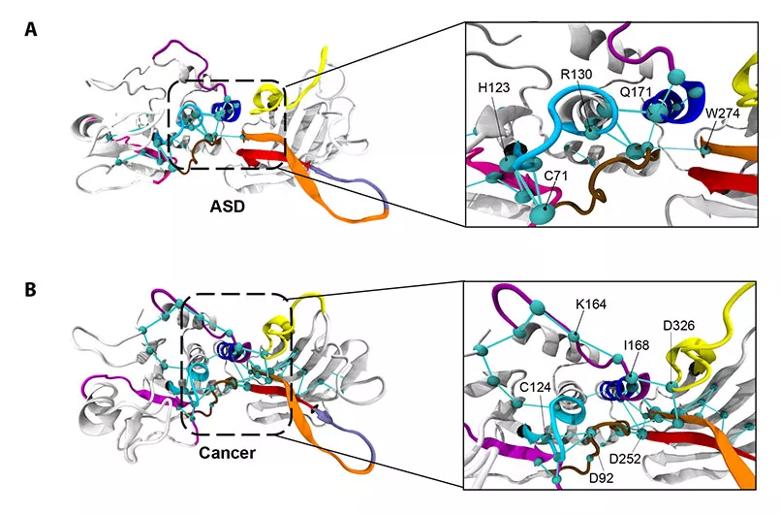
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/06a43c13-b11d-4cd2-8a4c-906acdae1322/805x-Inset-Lerner-Research-002_jpg)
The global metapaths of the ASD-associated mutant PTEN phenotype (top) and cancer-associated mutant PTEN phenotype (bottom).
Although patients with germline PTEN mutations present with a broad range of symptoms, they are universally diagnosed with PTEN hamartoma tumor syndrome (PHTS). Prior research from the Eng lab identified precise age-related penetrance of PHTS component cancers, the highest of which is breast cancer with an 85% lifetime risk. At an individual level, however, it is not currently possible to predict whether a patient with a PTEN mutation will fall in the 85% that will develop breast cancer or the 15% that won’t. Similarly, while it is known that up to 23% of PHTS patients develop ASD, it is not known which 23%.
In response to this diagnostic need, the Eng laboratory is currently investigating how advanced 3-D modeling techniques can detect structural and functional differences among PTEN variants that may be indicative of different diseases and, thus, better inform precision care. Their findings, recently published in the American Journal of Human Genetics, show that ASD- and cancer-driving PTEN variants differentially affect the structure and function of PTEN’s active site.
When functioning properly, PTEN inhibits a signaling pathway that promotes cell growth, survival and proliferation, helping to halt the growth and spread of cancerous cells. Dr. Eng and her team found that PTEN’s active site — the region that binds, like a lock and key, to other molecules to catalyze reactions — looks structurally different in PTEN mutations that result in ASD compared to cancer. As such, they bind differently and set off different reactions and signaling cascades, helping to explain why patients with variants to the same gene can present with vastly different diseases.
Advertisement
These findings are important for two reasons. As Dr. Eng explains, “We found that through sophisticated 3-D modeling, which collates complex network and molecular dynamics that we’ve never before collected, we can discern nuanced but important structural differences between germline PTEN variants that are associated with and predictive of a patient’s clinical condition. Our hope is that one day these techniques can be used in the clinic to directly inform a patient’s care and deliver a more accurate and meaningful diagnosis — either PTEN-ASD or PTEN-cancer, rather than the general PHTS.”
Additionally, this study opens the door for future investigations into targeted therapies. “Since we see that PTEN’s active site is structurally affected by mutations to the PTEN gene, there is reason to believe that targeting alternate binding sites, called allosteric regions, may be a viable, and currently unexplored, treatment approach for PTEN-cancer.” While more studies are needed, Dr. Eng is excited to explore PTEN’s potential allosteric druggability.
Iris Nira Smith, PhD, a T32 fellow, was first author on this study, which was funded in part by the Ambrose Monell Foundation, National Cancer Institute and National Institute of Neurological Disorders and Stroke.
Dr. Eng is the inaugural Chair of the Genomic Medicine Institute, inaugural Director of the Center for Personalized Genetic Healthcare and Medical Director of the PTEN Multidisciplinary Clinic. Notably, she was the first to discover a link between mutations in PTEN and Cowden and other syndromes, and her research has formed the basis of national practice guidelines for those with PHTS and Cowden syndrome. Dr. Eng holds the Sondra J. and Stephen R. Hardis Endowed Chair in Cancer Genomic Medicine at Cleveland Clinic.
Advertisement
Advertisement
First full characterization of kidney microbiome unlocks potential to prevent kidney stones
Researchers identify potential path to retaining chemo sensitivity
Large-scale joint study links elevated TMAO blood levels and chronic kidney disease risk over time
Investigators are developing a deep learning model to predict health outcomes in ICUs.
Preclinical work promises large-scale data with minimal bias to inform development of clinical tests
Cleveland Clinic researchers pursue answers on basic science and clinical fronts
Study suggests sex-specific pathways show potential for sex-specific therapeutic approaches
Cleveland Clinic launches Quantum Innovation Catalyzer Program to help start-up companies access advanced research technology