Pairing machine learning with multi-omics revealed potential therapeutic targets
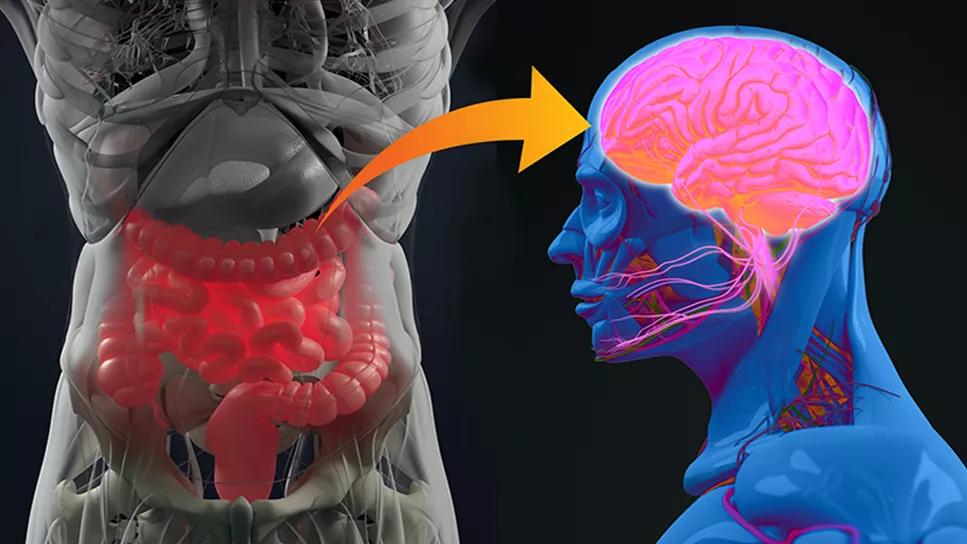
Image content: This image is available to view online.
View image online (https://assets.clevelandclinic.org/transform/769032c2-3042-4b96-a23b-fa2086c041dc/gut-brain-axis)
gut-brain axis
Using a machine learning approach, Cleveland Clinic researchers have discovered multiple potential therapeutic targets in Alzheimer’s disease (AD) by studying interactions between gut microbial metabolites and cellular receptors, some of which were previously unknown. Their work was recently published in Cell Reports (2024;43[5]:114128).
Advertisement
Cleveland Clinic is a non-profit academic medical center. Advertising on our site helps support our mission. We do not endorse non-Cleveland Clinic products or services. Policy
The gut-brain axis is an increasing area of focus in AD, with evidence of links between changes in the gut microbiome and AD pathophysiology in the central and peripheral nervous systems. However, the molecular mechanisms that could help explain the apparent AD/microbiome/metabolite connection are poorly understood.
The Cleveland Clinic researchers’ approach — which used an artificial intelligence (AI)-powered method to identify and evaluate more than a million gut metabolite/receptor protein pairs — found potentially druggable targets, particularly among pairs involving “orphan” G-protein-coupled receptors (GPCRs) whose physiological roles hadn’t been known.
Lab experiments verified that metabolite agonists of AD-associated GPCRs highlighted by the researchers’ computational analysis significantly reduced pathologically phosphorylated tau protein in stem cell-derived neurons from patients with AD.
The researchers’ approach is a “powerful tool” to identify possible AD therapies and could also generate lead compounds in other complex diseases, according to Feixiong Cheng, PhD, inaugural director of the Cleveland Clinic Genome Center and the study’s principal investigator.
“Gut metabolites are the key to many physiological processes in our bodies, and for every key there is a lock for human health and disease,” Dr. Cheng notes. “The problem is that we have tens of thousands of receptors and thousands of metabolites in our system, so manually figuring out which key goes into which lock has been slow and costly. That’s why we decided to use AI.”
Advertisement
Specifically, the team developed a systems biology framework that combined machine learning and multi-omics to analyze over 1.09 million potential metabolite/protein pairs among 335 gut microbial metabolites and 408 human GPCRs. The aim was to predict the likelihood that each interaction contributed to AD. The analyses integrated:
The analyses indicated potential functional roles of GPCRs — particularly orphan GPCRs — in AD pathophysiology, suggesting that orphan GPCRs could be potential drug targets. The researchers used multi-omics evidence from these analyses to identify GPCRs that appeared to be most associated with AD and to predict which metabolites had affinity for these receptors.
These efforts led them to focus on a protective metabolite called agmatine, believed to shield brain cells from inflammation and associated damage. The study found agmatine was most likely to interact with an orphan GPCR called C3AR in the setting of AD.
The researchers then found that C3AR levels were directly reduced when iPSC-derived neurons from AD subjects were treated with agmatine, demonstrating that metabolite and receptor influence each other. Additionally, AD neurons treated with agmatine also had lower levels of phosphorylated tau proteins, a marker for AD.
Similarly, another gut metabolite, phenethylamine, was found to significantly reduce tau hyperphosphorylation in iPSC-derived neurons from AD patients.
Advertisement
Dr. Cheng says these experiments demonstrate how his team’s AI algorithms can pave the way for new research avenues into diseases beyond AD. “We specifically focused on Alzheimer’s disease, but metabolite/receptor interactions play a role in almost every disease that involves gut microbes,” he notes. “We hope our methods can provide a framework to advance the entire field of metabolite-associated diseases and human health.”
Now, Dr. Cheng and his team are further developing and applying these AI technologies to study interactions between genetic and environmental factors (including food and gut metabolites) on human health and diseases, including AD and other complex disorders.
The present research involved collaboration between the Genome Center, part of Cleveland Clinic Lerner Research Institute, and Cleveland Clinic Luo Ruvo Center for Brain Health and the Center for Microbiome & Human Health (CMHH), also part of Lerner Research Institute. Study first author and Cheng lab postdoctoral fellow Yunguang Qiu, PhD, spearheaded a team that included J. Mark Brown, PhD, Director of Research, CMMH; James Leverenz, MD, Director of the Luo Ruvo Center for Brain Health and Director of the Cleveland Alzheimer’s Disease Research Center; and neuropsychologist Jessica Caldwell, PhD, ABPP/CN, Director of the Women’s Alzheimer’s Movement Prevention Center at Cleveland Clinic.
Advertisement
Advertisement
An expert talks through the benefits, limits and unresolved questions of an evolving technology
How we’re efficiently educating patients and care partners about treatment goals, logistics, risks and benefits
Large NIH-funded investigation is exploring this understudied phenomenon
Observational evidence of neuroprotection with GLP-1 receptor agonists and SGLT-2 inhibitors
Genomic study lays groundwork for insights into potential biomarkers and therapeutic strategies
Proteins related to altered immune response are potential biomarkers of the rare AD variant
Alzheimer’s studies delve into sex-related variances in the expression of the disease
Validated scale provides a method for understanding how lifestyle may protect against Alzheimer's